Course overview: This five unit lecture-laboratory course is a “capstone” course for cell biology majors (and anyone else who wants this experience). The goal is to provide you an opportunity to integrate the facts you’ve learned from the upper division BIS series into a more complete picture of how complex biological systems work and are studied (i.e., cells). This is best done by applying the logic of experimental design to addressing specific hypotheses associated with basic cellular functions. Though you may not use these exact methods in the future, it is a chance for you to participate in the techniques you’ve heard described in your courses and to practice using the logic of experimental design and trouble-shooting. The logic of design and trouble-shooting are invaluable skills that will serve you well in ANY career!
Access to “live” research questions: Every year since I began to teach this course, we have focused on a unique aspect of cell biology derived from either studies on mitochondria from the Nunnari Lab at UC Davis or more recently on studies of septins and cell division from my lab. Translating original cell biology questions from research labs to the teaching labs provides students with a realistic appreciation of the challenges and excitement of research. The questions that are brought to 140L derive from experiments we have done in my lab or from outstanding questions in the field. While the work of 140L students is necessarily a bit rushed due to the shortness of the quarter, I’ve been thrilled that every year students make unique observations. These new findings form seeds for UG research projects in my lab and for future generations of 140L experiments. Here is link to a recent MCB140L syllabus.
Unique observations and new projects: Below, in reverse chronological order, I will summarize the most exciting observations from each 140L class that has studied septins.
Techniques used in MCB140L:
Yeast transformation
Directed yeast two hybrid
Cell fractionation
SDS-PAGE and Western blot analysis using fluorescence
Membrane protein topology mapping
Affinity purification of protein complexes
Database and bioinformatic analyses
Fluorescence microscopy using GFP fusions
Quantitative analysis of fluorescence using Image J
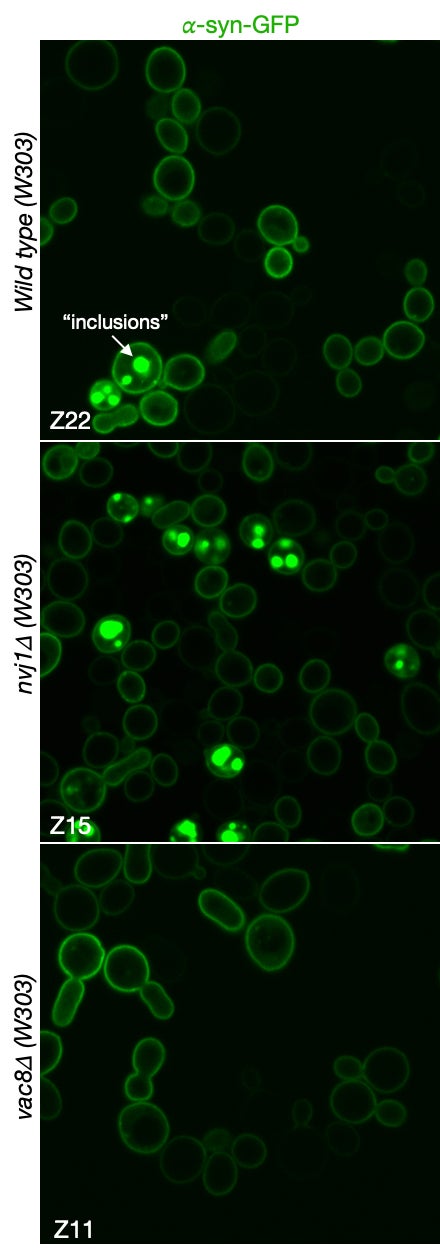
MCB140L 2022-23: After the disruption of the COVID years (2021 and part of 2022), I designed MCB140L around a related but novel question: how do changes in cellular autophagy pathways affect the accumulation of protein aggregates, inclusions and organelle damage associated with neurodegenerative diseases? This has been an exciting new focus both in my own research lab but also for 140Lers. The major gap in understanding N.D. onset and progression centers around the proposed causal role of protein aggregates and inclusions. The primary evidence is the strong correlation between protein inclusions in patients’ brains post-mortem and neurological symptoms of dementia, a causal relationship supported by studies in animal models. However, the formation of such inclusions does not shed light on the pathways that are affected in cells that result in toxicity and loss of neuronal function. The MCB140L lab used budding yeast to probe the relationship between autophagy, a pathway proposed to protect against N.D., and the formation of inclusions and the increase in cellular toxicity observed when human alpha-synuclein is over-expressed. Interestingly, students experiments suggested that loss of a very specific microautophagy pathway conferred increased cellular toxicity, while failing to increase inclusions. This finding is very exciting as it calls into question the simple relationship between protein inclusions and cellular toxicity, raising the possibility that toxicity originates earlier in the N.D. process through perturbations of cellular stress response pathways. The images below demonstrate this remarkable relationship. The images show a-synuclein fused to GFP and expressed in wild type, nvj1d and vac8d cell lines. The NVJ1 gene encodes for a protein that links the nucleus and nuclear ER to the vacuole through an interaction with protein encoded by the VAC8 gene. Interestingly, Vac8 also helps to localize autophagosomes that form in response to cell stress. The formation of inclusions in wild type and nvj1d cells are clearly evident (see arrow in wild type cells) but are strikingly absent from vac8d cells. On the surface, this suggests that autophagosome biogenesis is linked to inclusion formation. This observation awaits further validation and additional experience to allow us to better understand the relationship between autophagy and N.D. Come join us in MCB140L or my lab to discover more about this very interesting finding.
MCB140L 2019-2020: The lab class has built on the observations from 2018 and work in my lab to probe how septins impact the trafficking of autophagy specific membranes. In 2020 the lab focused on measuring autophagy using fluorescence imaging and quantitative approaches that helped to confirm that cells carrying mutations in septin genes exhibit an increase in autophagy!
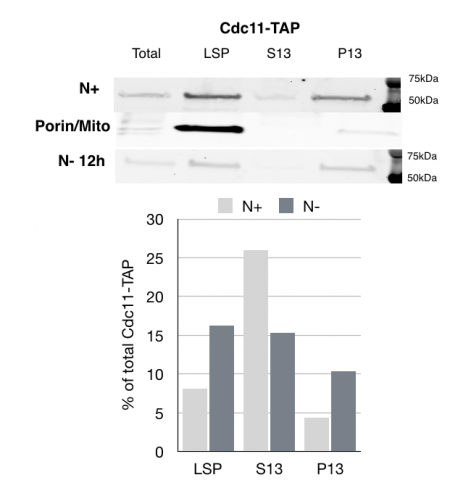
MCB 140L 2018, Septins role in autophagy: Based on Jonathan Mendes work and follow up work in my lab on septins reorganization after nutrient deprivation, this year’s students examined the impact of starvation on septins. One of the major findings made my students is that septins fractionate with larger membranes after nutrient deprivation. The class of membranes are not entirely clear from the experiments would could carry out, but these results suggest that there is a clear transition in the way septins associate with cellular membranes after starvation induced autophagy.
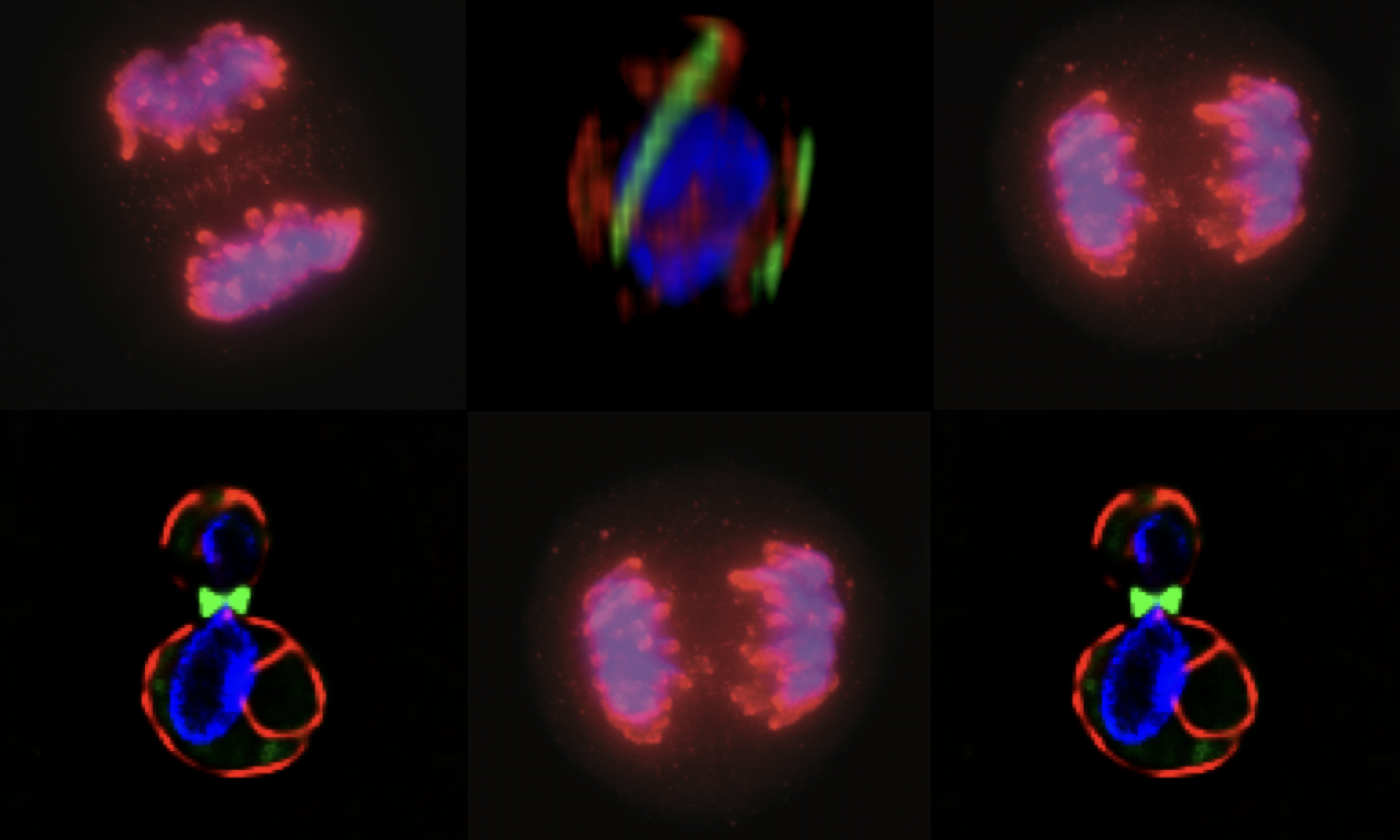
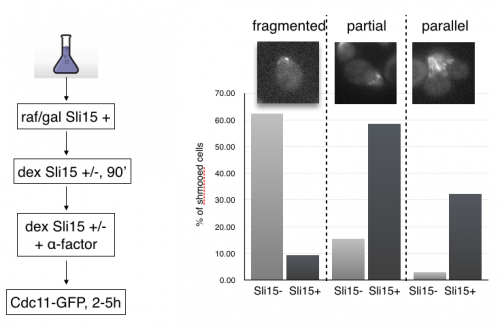
MCB140L 2016: CPC regulation of septins during the mating program in budding yeast: My lab had published a role for chromosome passenger complexes in regulating septin dynamics in anaphase. The 140Lers asked if these CPCs were also important for regulating septin dynamics in other circumstances. We focused on the cell polarity signal initiated by mating pheromone that results in the formation of a mating projection in budding yeast. This structure, known as a “shmoo” had not be characterized for changes in septin organization. Students beautifully showed that septins reoriented from an array of filaments parallel to the cell axis to a perpendicular arrangement after mating pheromone addition. Remarkably, they showed that CPCs are required for this transition!! This is the first evidence that CPC have a role outside of mitosis in regulating septins. In analyzing septin behavior under different growth conditions, one 140Ler (Jonathan Mendes) noticed that septins had fragmented in cells that were “stressed”. Jonathan formulated a hypothesis that these fragmented septins might be associated with membranes involved in mitophagy. He came to my lab the following summer and launched an entirely new set of studies on septins and autophagy (the process by which cells “eat” their own organelles to survive stress conditions like starvation). This Is a great example of how a student’s observations and imaginative leap can drive research forward.
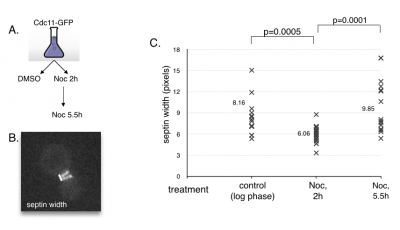
MCB140L2014: Cell cycle advance and septin dynamics. My lab had filmed septin GFP fusions and noted that septin rings become distinct as cells entered anaphase. Students tested the hypothesis that septin ring formation is under control of cell cycle regulators. First, they tested the behavior of septin rings upon arresting cells in pre-anaphase using a drug that engages the spindle assembly checkpoint. The prediction is that septins will remain in a collar like organization when arrested. Second, they repeated this experiment in a strain that contained a mutation in the spindle assembly checkpoint. This prediction here was since the cells would not arrest in the presence of the drug, septin rings should separate. Students found that septin rings formed and separated even when arrested prior to anaphase onset. They also found that the absence of the spindle checkpoint resulted in septin rings failing to fully separate before exit from mitosis. These results suggest that septin ring formation and separation is not linked to the cell cycle regulators and that it may rely more on insertion of cell membranes and thus cell growth.